| Metagenome Orchestramago | |
| |
|
 |
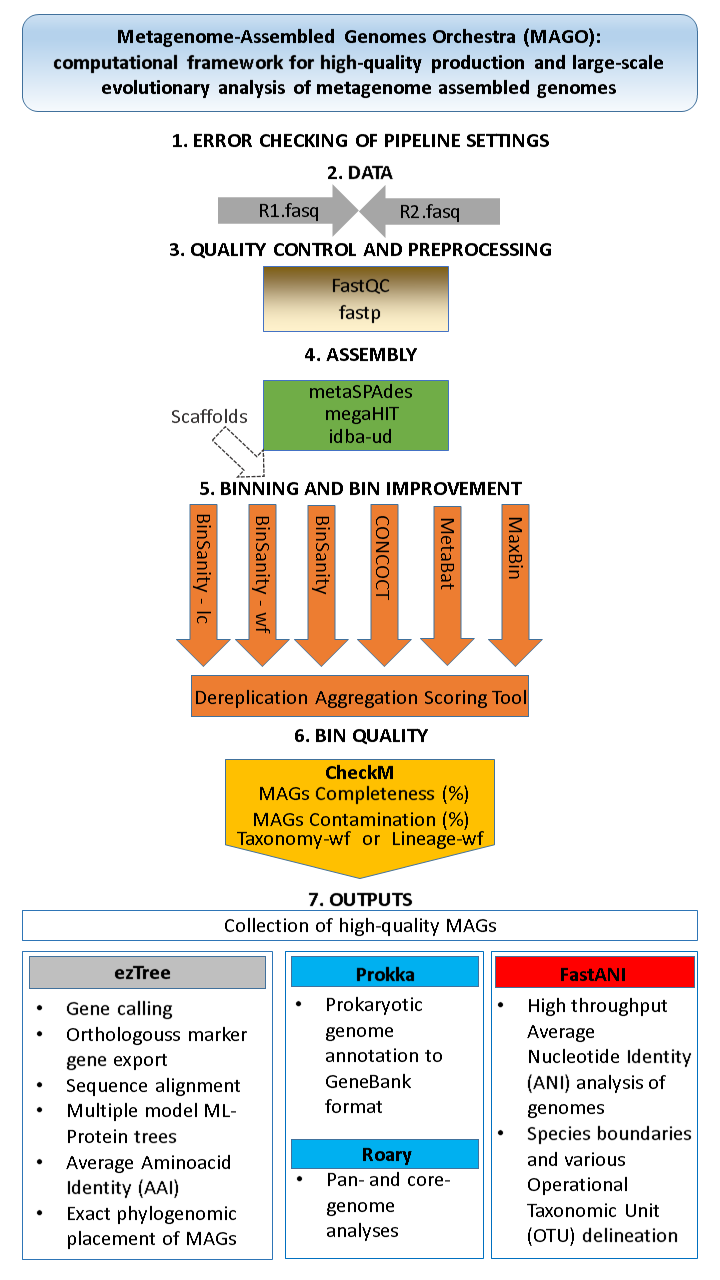
• user-friendly out-of-the-box metagenome assembly, binning and evaluation |
| • suitable for autonomous and batch execution on High-Performance Computing (HPC) facilities | |
| • automatic crash recovery in case of interruption | |
| • memory usage limiting to avoid system destabilization due to excessive swapping | |
| • input reads evaluators: FastQC and FastP | |
| • assemblers: Idba-UD, MegaHIT and metaSPAdes | |
| • possibility of using externally built scaffolds | |
| • SAM/BAM file generation methods: Bwa, Bowtie2 and BBMap | |
| • binners: MaxBin, Concoct, MetaBat2, BinSanity (plain, workflow and LC) | |
| • refinement of bins with DasTool | |
| • evaluators and processors: CheckM, ezTree, Prokka, Roary and FastANI | |
| • all contained software ingredients preinstalled and preconfigured | |
| • automatic handling of intermediate files and their formats | |
| • automatic handling of command-line parameters | |
| • experienced users can freely pass their own command-line parameters to underlying applications | |
| Licensing: Creative Commons Attribution CC BY licence |
|
IMPORTANT.
Metagenome Orchestra is a skeleton application for a synergic
execution of many externally developed pieces of software.
These are disseminated as integrated parts of Orchestra to
provide the level of end-user experience that the system aims
to deliver. Nonetheless, every included piece of software
remains owned and copyrighted by its respective developers.
Please see document credits.txt for a list of included software. |
| IMPORTANT. Metagenome Orchestra is developed and disseminated in a good faith and desire to work according to expectations, but authors DO NOT give any guarantees about its correctness. USE IT AT YOUR OWN RISK. Authors cannot be held legally or morally responsible for any consequences that may arise from using or misusing Orchestra. |
| Reference for citation |
| If you use Metagenome Orchestra, please cite the following reference. |
|
Murovec, Bostjan & Deutsch, Leon & Stres, Blaz. (2019). Metagenome-Assembled Genomes Orchestra (MAGO): computational framework for high-quality production and large-scale evolutionary analysis of metagenome assembled genomes. Molecular biology and evolution. 37. 10.1093/molbev/msz237.
|
|
Please also cite the included software that you use as part of your pipeline processing. Please see document credits.txt for a list of included software. |
| Download links |
| Metagenome Orchestra is available as a Singularity image, Docker image or Oracle VirtualBox virtual machine. |
| The most recent version V2.2b was released on 8. 3. 2020 as a singularity image only (list of changes). Version in Docker container and Virtual Machine is V2.2a, which was released on 4. 9. 2019. |
| Download link for Metagenome Orchestra Singulariy image (4.2 GB). |
| Download link for Metagenome Orchestra Docker image (7.1 GB). |
| Download link for Metagenome Orchestra Virtual Machine (6.6 GB). |
| Optionally, advanced users may want to check integrity of the above Orchestra containers by examining their SHA256 hashes, which are calculated with the sha256sum utility. |
| Generally, the easiest way to start using Orchestra and to get familiar with it is to run it in a Virtual Box Machine. However, this option is less suitable for running large-scale analyses on high-performance computing facilities. |
|
First impressions about Metagenome Orchestra can be obtained by running a small demo
analysis available as a ZIP archive (0.3 GB).
Please follow the below platform-specific instructions, which are also included in the demo archive.
Note: the Virtual Box release of Metagenome Orchestra already contains the demo. You can run it by clicking a desktop icon within the virtual machine. |
|
• For Singularity image: singularity_instructions.txt • For Docker container: docker_instructions.txt • For VirtualBox virtual machine: virtualbox_instructions.txt |
| The procedure for running your own real metagenome analyses is as follows. |
| 1. Prepare configuration file according to template file config_template.txt, which is self descriptive and also serves as a platform agnostic user manual. It contains a detailed description of processing features and Orchestra pipeline. |
| 2. Take into account platform-specific instructions about specification of file names and running Orchestra. These are included in the respective versions of instructions above. |
| FastANI corner | |
|
In order to successfuly utilize the
FastANI part of Orchestra processing
it is necessary to follow the FastANI instructions
and provide Orcherstra an appropriate external database for genome comparison.
File or directory with external database is specified through configuration parameters as described in template file config_template.txt. This can be either a collection of draft genomes produced within ones own study, an external narrow database for analyses of single specific phylogenetic group (for examples databases from D1 to D5 at http://enve-omics.ce.gatech.edu/data/fastani), or a general database containing a large number of high quality genomes (for example the NCBI database at the same link http://enve-omics.ce.gatech.edu/data/fastani) for assessing species boundaries of novel cohort of draft metagenome assembled genomes. Genome Taxonomy Database The users are also directed to consider Orchestra as the first step (taking the raw sequencing data to metagenome assembled genomes) to an emerging platform of Genome Taxonomy Database (https://gtdb.ecogenomic.org) and provides ongoing improvements in exact prokaryotic genome taxonomy. |
| Contact | |
| We look forward to receive your feedback regarding the use of Metagenome Orchestra, potentially discovered bugs and issues, as well as comments and suggestions for further development of the system. | |
|
Blaž Stres (metagenomic related topics)
<mailto:blaz.stres@bf.uni-lj.si> University of Ljubljana, Biotechnical faculty, Slovenia University of Ljubljana, Faculty of Medicine, Slovenia UNI-LJ, Faculty of Civil and Geodetic Engineering, Slovenia Jozef Štefan Institute, Ljubljana, Slovenia Institute of Microbiology, University of Innsbruck, Austria |
Boštjan Murovec (computing related topics)
<mailto:bostjan.murovec@fe.uni-lj.si> University of Ljubljana, Faculty of electrical engineering, Slovenia Leon Deutsch <mailto:leon.deutsch@bf.uni-lj.si> University of Ljubljana, Biotechnical faculty, Slovenia |